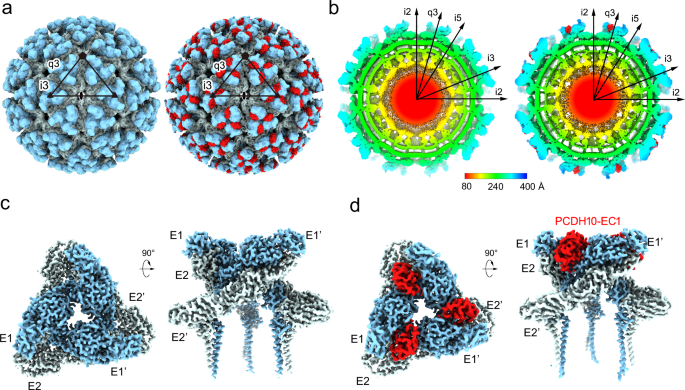
Cryo-EM structure determination
The WEEV strain 71V1658 VLPs and human PCDH10-EC1-EC2 fused with a C-terminal human antibody constant region (Fc) tag were produced in HEK293F cells (Supplementary Fig. 1). A total of 3106 or 17,711 cryo-electron micrographs of WEEV VLPs alone or in complex with PCDH10-EC1-EC2-Fc, respectively, were used for further reconstruction (Supplementary Fig. 2 and Supplementary Table 1). Single-particle reconstructions with icosahedral symmetry yielded a nominal resolution of 4.50 Å or 5.90 Å with 40,222 or 17,858 good icosahedral particles for WEEV VLPs alone or in complex with PCDH10-EC1-EC2-Fc, respectively (Fig. 1a, b and Supplementary Fig. 2). To improve the densities, symmetry extension, block-based classification, and local refinement focusing on the q3 spikes were applied to achieve a final resolution of 2.53 or 2.99 Å for WEEV VLPs alone or in complex with PCDH10-EC1-EC2-Fc, respectively (Methods; Fig. 1c, d, Supplementary Figs. 2 and 3). By using the cryo-EM densities as a guide, residues 1-96 of PCDH10-EC1 in the complex were built. The models of WEEV E1 (residues 1–439) and E2 (residues 5–419) were also built in the densities. In contrast, the residues of PCDH10-EC2 and the fused Fc tag were not visible in the densities, likely due to a lack of interaction with WEEV VLPs, leading to a flexible conformation. Given that recombinant PCDH10-EC1-EC2 alone exhibited poor stability, we fused PCDH10-EC1-EC2 with an Fc tag through a flexible linker to optimize its biochemical properties, which is a widely used strategy in alphavirus receptor studies25,30,31,38. The lack of density corresponding to the Fc tag indicates that the fusion of PCDH10-EC1-EC2 with the Fc tag through a flexible linker would not impact the WEEV:PCDH10-EC1-EC2 interaction in structural biology and biochemical studies.
a The colored surface representations of cryo-EM maps of WEEV VLPs alone (left panel) and in complex with PCDH10-EC1-EC2 (right panel). The densities of WEEV E1 and E2 proteins are colored pale cyan and light blue, whereas the densities of PCDH10-EC1 are colored red. The 5-fold, 3-fold, and 2-fold icosahedral axes are indicated by a pentagon, triangles, and an oval, respectively. The i3 spike and q3 spike are indicated by the labels “i3” and “q3” respectively. b The equatorial cross-sections of WEEV VLPs alone (left panel) and in complex with PCDH10-EC1-EC2 (right panel) in radial distance color scheme. The radial distances are indicated by the bar. The density of PCDH10-EC1 is separately colored red for highlighting in the right panel. c, dCryo-EM maps achieved by the final refinement focusing on the q3 spike in WEEV VLPs alone (c) or in complex with PCDH10-EC1-EC2 (d). The densities are shown as a colored surface in two perpendicular views respectively.
Overall structure
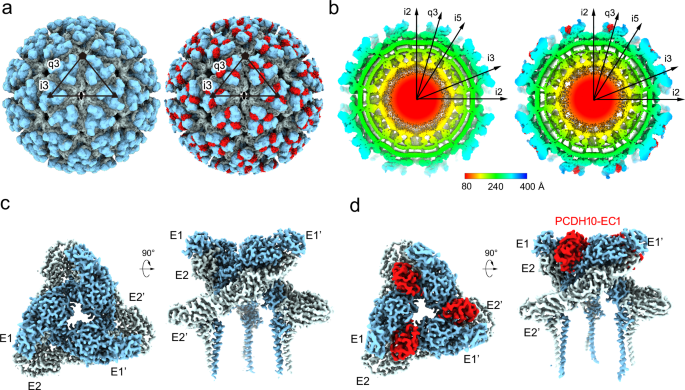
In the overall density of the complex, a total of 240 receptor molecules bind to WEEV VLPs, and the densities of PCDH10-EC1 present an identical binding mode at the q3 spikes and the i3 spikes (Fig. 1a). PCDH10-EC1 inserts into a cleft clamped by two adjacent E2-E1 heterodimers in the trimeric spike, where it forms contacts with E1 and E2 in one E2-E1 heterodimer (named E2-E1) and E2’ in an adjacent heterodimer (named E1’-E2’) (Fig. 2a, b). The contact surface area of PCDH10-EC1 was calculated to be 1080 Ų, which is a significant portion of its total accessible surface area of 6,216 Ų, indicating a strong virus‒receptor interaction. The contacting areas on PCDH10-EC1 accounting for the interactions with E1, E2, and E2’ were calculated as 235 Å2, 377 Å2 and 468 Å2, respectively.
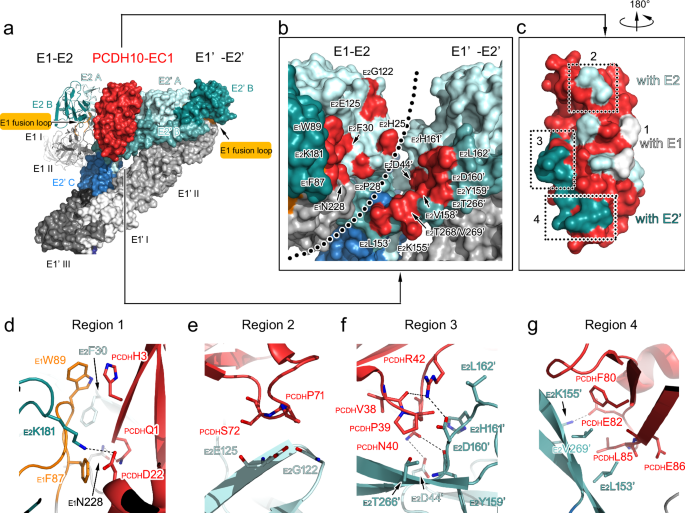
a–c Structures of two WEEV E2-E1 heterodimers in complex with a PCDH10-EC1. aOne E2-E1 heterodimer is shown as a colored cartoon, while the adjacent heterodimer (E1’-E2’) is covered by a molecular surface. The color scheme for E1/E2 domains and PCDH10-EC1 is as following: E1_I, white; E1_II, light gray; E1 fusion loop, orange; E1_III, gray; E1_ecto and E1_TM, deep gray; E2_A, pale cyan; E2_β, light teal; E2_B, deep teal; E2_C, sky blue; E2_TM, deep blue; PCDH10-EC1, red. b The E1/E2 residues that interact with PCDH10-EC1 are colored in red on the molecular surface with labels in an enlarged view. Two E2-E1 heterodimers are both covered by molecular surfaces. The boundary of two E2-E1 heterodimers is roughly indicated by a dotted line. c PCDH10-EC1 is covered by a molecular surface with its interacting residues for contact with E1, E2, and E2’ in white, pale cyan, and light teal, respectively. The interaction regions 2-4 are indicated by dashed frames. d–g Details for the virus-receptor interaction. The interacting regions 1 (d), 2 (e), 3 (f), and 4 (g) are presented in enlarged views. All polypeptides are shown as cartoon diagrams. Interacting residues are displayed as colored sticks with labels. The subscripts of the labels indicate the proteins that the residues belong to. Dashed lines denote the hydrogen bonds or salt bridges. For note, some interactions are not involved in these panels due to clear views and representations.
Upon binding with PCDH10-EC1, the E1 and E2 proteins of WEEV VLPs exhibit local conformational changes at the interfaces (Supplementary Fig. 4a, b). Similarly, PCDH10-EC1 also shows conformational changes in the interacting region, particularly in a loop region boundaried by a disulfide bond formed by C67-C73 (Supplementary Fig. 4c). The conformations of other portions of WEEV E1/E2 and PCDH10-EC1 were not altered. Two calcium ions were identified in the crystal structure of PCDH10-EC1 (PDB: 6VG433), and the side chains of their interacting residues did not show convincing density (Supplementary Fig. 4c, d). Moreover, the calcium ions are positioned remotely from the WEEV VLP interface, suggesting that the binding is likely independent of calcium ions.
Interactions of WEEV VLPs and PCDH10-EC1
The interacting residues of PCDH10-EC1 include residues PCDHQ1 and PCDHH3, which interact with E1; residues PCDHQ1, PCDHD22, PCDHP71, PCDHS72, PCDHH76 and PCDHQ89, which interact with E2; and residues PCDHV38, PCDHP39, PCDHN40, PCDHR42, PCDHF80, PCDHE82, PCDHL85 and PCDHE86 on PCDH10-EC1, which interact with E2’ in the adjacent E1’-E2’ heterodimer (Fig. 2b, c and Supplementary Table 2). These residues are distributed in four main regions (regions 1–4) (Fig. 2c).
Region 1 contains the PCDH10-EC1 residues mainly in the central β-sheet (PCDHQ1, PCDHH3, PCDHD22 and PCDHQ89), which interact with the residues of E1 (E1F87, E1W89 and E1N228) and E2 (E2F30 and E2K181) of WEEV VLPs (Fig. 2d). In this region, the side chain of the PCDHQ1 residue forms a hydrogen bond with the side chain of E1N228. The side chains of residue E1F87 in the E1 fusion loop and residue E2F30 also offer close contact with the main-chain atoms of residue PCDHQ1. Moreover, the large side chain of residue E1W89 located in the E1 fusion loop stacks with the side chain of PCDHH3. A hydrogen bond formed by the side chain of PCDHD22 of PCDH10-EC1 and E2K181 of E2 was also identified in this region.
In region 2, residues PCDHP71-PCDHS72 of PCDH10-EC1 face E2G122 and E2E125 of E2, forming close contacts and stabilizing the interaction of PCDH10-EC1 with WEEV VLPs (Fig. 2e). The closest intermolecular distance among the interactions in this region is ~ 3.2 Å, with the distances for other interactions ranging from 3.5–4.0 Å, indicating a minor role of this region in virus–receptor recognition (Supplementary Table 3).
The interaction network in region 3 consists of a loop region spanning residues PCDHV38-PCDHR42 of PCDH10-EC1 and residues of E2’ (including E2D44’, E2V158’, E2D160’, E2H161’, E2L162’ and E2T266’) (Fig. 2f). The main-chain atom of PCDHV38 interacts with the side chain of PCDHR42, stabilizing the conformation of PCDHR42 together with an interaction with the side chain of E2D160’. Residue PCDHP39 faces E2V158’, E2D160’, E2H161’, and E2T266’, with the distances of the contacts ranging from 3.4–4.0 Å. The side chains of E2D160’ and E2H161’ of E2’ are close to PCDHN40 and PCDHR42, additionally contributing to the receptor‒virus interaction.
In region 4, the side chains of PCDHF80 and PCDHL85 are hydrophobically packed against the side chains of E2L153’ and E2V269’ (Fig. 2g). Moreover, the side chain of PCDHE82 forms a hydrogen bond with the side chain of E2K155’. The side chain atoms of E2L153’ are also in contact with the main-chain atoms of PCDHL85 and PCDHE86, playing an additional role in the interaction.
To validate whether calcium ions are required for the interaction between WEEV VLPs and PCDH10-EC1-EC2-Fc, we conducted BLI assays under calcium depletion conditions using a range of EGTA concentrations from 0 to 25.0 mM, our results showed that even at concentrations as high as 25.0 mM EGTA, the formation of the WEEV VLP:PCDH10-EC1-EC2-Fc complex was not significantly inhibited, confirming that calcium ions are not required for the interaction, consistent with the structural observations (Fig. 3a).
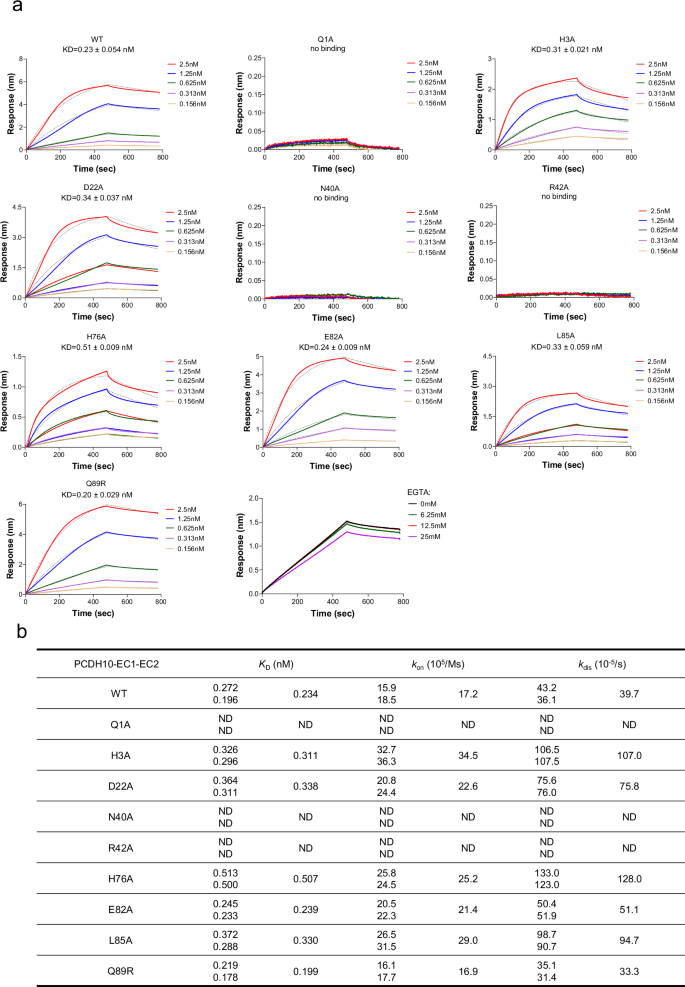
a The binding and disassociation curves of WT PCDH10-EC1-EC2 and a representative mutant PCDH10-Q1A, -D22A, -N40A, -R42A, -H76A, -E82A, -L85A, and -Q89R with WEEV 71V1658 VLPs and WT PCDH10-EC1-EC2 with WEEV 71V1658 VLPs with 0, 6.25, 12.5, 25.0 mM EGTA are shown. The calculated KD values are displayed in each corresponding panel. The displayed results were obtained from a single technical replicate and are representative of two biologically independent experiments. The equilibrium dissociation constant (KD) was calculated as the mean (n = 2). b Summary of the measured binding affinities. The KD value for each sample is the mean value of two independent measurements. kon, association rate constant; kdis, dissociation rate constant; ND, too weak interaction cannot be detected.
Mutagenesis studies of key interacting residues of PCDH10-EC1
To assess the impacts of interacting residues on virus‒receptor engagement, we mutated a set of interacting residues from PCDH10-EC1-EC2-Fc and measured their binding affinities with WEEV VLPs via biolayer interferometry (BLI) (Fig. 3a). As a positive control, the wild-type (WT) PCDH10-EC1-EC2-Fc binds to WEEV VLPs with an affinity constant (KD) of 0.234 nM. The KD values reflect the macroscopic binding affinities between PCDH10-EC1-EC2-Fc and WEEV VLPs, whereas Kon and Koff represent the association rate constants and dissociation rate constants, respectively. The KD values obtained from BLI act as relative indicators of binding strength and show trends in affinity variations between different proteins and WEEV VLPs. However, they only offer a rough estimate of the true KD values.
Among all the selected interacting residues, PCDHQ1, PCDHN40, and PCDHR42 form strong bonds with WEEV residues in regions 1 and 3. Substitution of these residues with alanine residues eliminates the ability to bind with WEEV VLPs, which is consistent with their robust interactions with WEEV VLPs observed in the structure. To study the importance of PCDHQ1, PCDHN40, and PCDHR42 for WEEV infection, HEK293T cells stably expressing the indicated PCDH10 proteins were infected with SINV-WEEV (71-V1658) (MOI = 0.5) for 9 h, and the expression of the indicated proteins was confirmed by immunoblotting (Fig. 4c). The PCDHN40 substitution abolished the entry of SINV-WEEV, as did the PCDHR42 substitution (Fig. 4a, b). In addition, single-residue mutation of PCDHQ1 substantially attenuated the entry of the SINV-WEEV viruses (Fig. 4a, b). All these results acquired from cell-based assays are consistent with the binding affinities measured by BLI, indicating the key roles of PCDHQ1, PCDHN40, and PCDHR42 in WEEV infection.
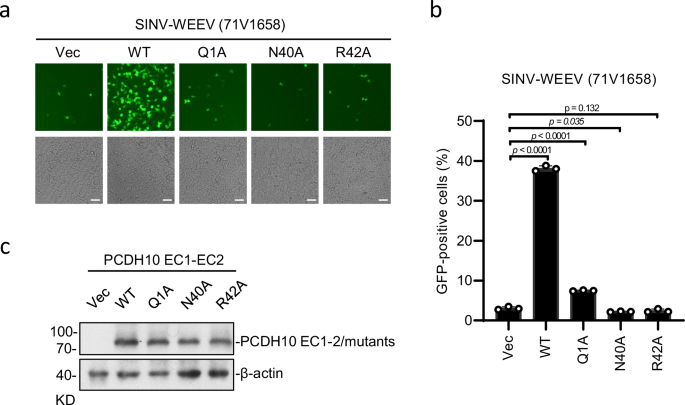
HEK293T cells stably expressing indicated PCDH10 proteins were infected with SINV-WEEV (71V1658) (MOI = 0.5) for 9 h. Infection was monitored by fluorescence microscopy (a) or flow cytometry (b). Expression of the indicated proteins was confirmed by immunoblots (c). Scale bars, 50 µm (a). Data are mean ± SD from 3 biological repeats, and p-values are from an unpaired two-tailed Student’s t test (b). At least three independent experiments were performed with similar results, and representative images are shown (a, c). Source data are available as a Source Data file.
The PCDHH3 residue is packed against E1W89 at a distance of ~ 3.6 Å, and the side chain of PCDHD22 forms a salt bridge with E2K181 at a distance of ~ 3.2 Å. The mutations of either of these residues (PCDH10-H3A and PCDH10-D22A) result in modestly decreased binding with WEEV VLPs, with KD values of 0.311 nM and 0.338 nM, respectively (Fig. 3a). The side chain of PCHDH76 is packed against the side chain of E2H25 at a distance of approximately 3.2 Å, and the disruption of this interaction by the PCDH10-H76A mutant decreased the KD (0.507 nM) compared with that of WT PCHD10-EC1-EC2. Residue PCDHL85 participates in hydrophobic interactions with E2L153’, and mutation of this residue (PCDH10-L85A) resulted in a modestly attenuated KD value of 0.330 nM. Notably, the dissociation rates for the interaction between WEEV VLPs and the PCDH10-H3A, PCDH10-D22A, PCDH10-H76A, and PCDH10-L85A mutants increased 1.9–3.2-fold, indicating that these complexes are more unstable. The PCDH10-E82A mutant presented a comparable KD value (0.239 nM) with WT PCDH10-EC1-EC2-Fc, suggesting that the hydrogen bond (~ 3.0 Å) formed by the side chains of PCDHE82 and E2K155’ plays a minor role in virus-receptor binding.







